把subset_data的子集重新放到总群allmerge中找marker基因 findallmarkers
把subset_data的子集重新放到总群allmerge中找marker基因 findallmarkers
Idents(All.merge)=All.merge$cell.type
levels(All.merge)
rownames(All.merge@meta.data[Idents(All.merge)=="Monocyte",])
All.merge@meta.data$new.cluster.idents=ifelse( Idents(All.merge)=="Monocyte","Monocyte",
ifelse( Idents(All.merge)=="T cell","T cell",
ifelse( Idents(All.merge)=="B cell","B cell",
ifelse( Idents(All.merge)=="Ig-producing B cell","Ig-producing B cell",
ifelse( Idents(All.merge)=="Dendritic cell","Dendritic cell",
ifelse( Idents(All.merge)=="Neutrophil","Neutrophil",
ifelse( Idents(All.merge)=="NK cell","NK cell",
ifelse(Idents(All.merge)=="Endothelial cell-1","Endothelial cell-1",
ifelse( Idents(All.merge)=="Endothelial cell-2","Endothelial cell-2",
ifelse( Idents(All.merge)=="Fibroblast","Fibroblast",
ifelse(Idents(All.merge)=="Myofibroblast/vascular smooth muscle cell","Myofibroblast/vascular smooth muscle cell",
ifelse(Idents(All.merge)=="Cycling basal cell","Cycling basal cell",
ifelse(Idents(All.merge)=="Ciliated cell","Ciliated cell",
ifelse(Idents(All.merge)=="Clara cell","Clara cell",
ifelse(Idents(All.merge)=="AT1 cell","AT1 cell",
ifelse(Idents(All.merge)=="AT2 cell-1","AT2 cell-1",
ifelse(Idents(All.merge)=="AT2 cell-2","AT2 cell-2",
ifelse(Idents(All.merge)=="Igha+ AT2 cell","Igha+ AT2 cell",
ifelse( rownames(All.merge@meta.data) %in% union(cellname_mycluster[["0"]],seq(1,ncol(All.merge)-length(cellname_mycluster[["0"]]),1) ),"cluster0",
ifelse(rownames(All.merge@meta.data) %in% union(cellname_mycluster[["1"]],seq(1,ncol(All.merge)-length(cellname_mycluster[["1"]]),1) ),"cluster1",
ifelse(rownames(All.merge@meta.data) %in% union(cellname_mycluster[["3"]],seq(1,ncol(All.merge)-length(cellname_mycluster[["3"]]),1) ),"cluster3","cluster.mixed4_5")
))
))))))))))))))))))
table(All.merge$new.cluster.idents)
table(All.merge$cell.type,All.merge$orig.ident)
Idents(All.merge)=All.merge$new.cluster.idents
markers=FindAllMarkers(All.merge,only.pos = TRUE,min.pct = 0.25)
接下来从findallmarkers得到marker基因,选特定cluster批量做dotplot气泡图
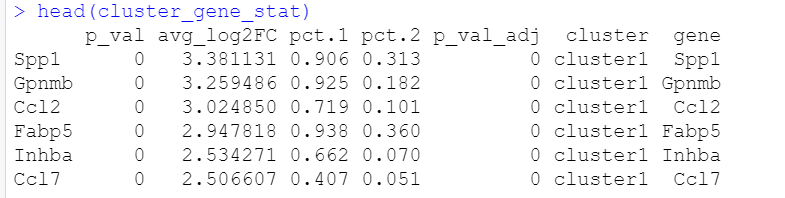
cluster_gene_stat=markers[markers$cluster %in% c("cluster1",
"cluster.mixed4_5", "cluster0","cluster3"), ]
head(cluster_gene_stat)

unique(cluster_gene_stat$cluster)
table(cluster_gene_stat$cluster)
cluster_marker=list()
for(i in unique(cluster_gene_stat$cluster)){ #按照p0.05值、flodchange 1标准 获得11个cluster的marker基因 列表
cluster_marker[[paste("cluster", i, sep = "_")]] <- cluster_gene_stat[cluster_gene_stat$avg_log2FC>1
&cluster_gene_stat$p_val<0.001
&cluster_gene_stat$cluster==i, "gene"]
}
head(cluster_marker)
unlist(lapply(cluster_marker, length))
getwd()
#批量做气泡图 Dotplot
for (j in names(cluster_marker)) {#一定要记得是向量 而不是一个数 for j in(1:11) 一定记得加括号!!!1:length(cluster_marker!!!!!!!!!
#j=5
times=(length(cluster_marker[[j]])+10)/10
times
if (floor(times)>=2 ){
for (i in seq(1,330,10)) { #使用循环出图 画图
#i=1
if ((i+9)>(10*floor(times))) {next}
print(cluster_marker[[j]][i:(i+9)])
jpeg(paste0("cluster_",j,"_",paste0(unlist(strsplit(cluster_marker[[j]][i:(i+2)], " "))[seq(1,3,1)],seq='_',collapse = "") , ".jpeg"),height = 9, width = 18, units = 'in', res=300)
p=DotPlot(All.merge,
features = cluster_marker[[j]][i:(i+9)])
print(p)
dev.off()
print(paste0("cluster_",j));print(cluster_marker[[j]][i:(i+9)])
}
print(j)
} else{ #条件一定记得加括号! !!!!!!!!!!!!!!!11!!!!!!!!!!!
print(j); print(cluster_marker[[j]][1:(1+9)])
jpeg(paste0("cluster_",j,"_",paste0(unlist(strsplit(cluster_marker[[j]][1:(1+2)], " "))[seq(1,3,1)],seq='_',collapse = "") , ".jpeg"),height = 9, width = 18, units = 'in', res=300)
p=DotPlot(All.merge,features = cluster_marker[[j]][1:(1+9)])
print(p)
dev.off()
}
}
#测试用
#以下 为踩坑教程 要么是等号的问题
要么是%in%谁在前谁在后的问题
要么是ifelse判断条件的问题
要么是union取并集的元素是否有重复的问题
if(1==1){#https://www.biostars.org/p/408922/
#AllCells.combined$IGHMPOS <- WhichCells(AllCells.combined, expression = Ighm > 0)
#https://github.com/satijalab/seurat/issues/2081
#https://www.datasciencemadesimple.com/union-in-r/
#https://github.com/satijalab/seurat/issues/2081
#https://github.com/satijalab/seurat/issues/3079
#测试用的
if(1==1){All.merge$newid=ifelse(cellname_mycluster[["0"]] %in% colnames(All.merge),"cluster0",
ifelse(cellname_mycluster[["1"]] %in% colnames(All.merge),"cluster1",
ifelse(cellname_mycluster[["3"]] %in% colnames(All.merge),"cluster3","mix4_5")))
All.merge$newid=ifelse(cellname_mycluster[["0"]] %in% colnames(All.merge),"cluster0",
ifelse(cellname_mycluster[["1"]] %in% colnames(All.merge),"cluster1",
ifelse(cellname_mycluster[["3"]] %in% colnames(All.merge),"cluster3","mix4_5")))
All.merge$newid= ifelse(match(cellname_mycluster[["0"]],colnames(All.merge)),"cluster0",
ifelse(match(cellname_mycluster[["1"]],colnames(All.merge)),"cluster1",
ifelse(match(cellname_mycluster[["3"]],colnames(All.merge)),"cluster3","cluster.mixed4_5")))
All.merge$newid= ifelse(colnames(All.merge)[match(cellname_mycluster[["0"]],colnames(All.merge))],"cluster0",
ifelse(colnames(All.merge)[match(cellname_mycluster[["1"]],colnames(All.merge))],"cluster1",
ifelse(colnames(All.merge)[match(cellname_mycluster[["3"]],colnames(All.merge))],"cluster3","cluster.mixed4_5")))
All.merge$newid= ifelse(cellname_mycluster[["0"]] %in% names(Idents(All.merge)),"cluster0",
ifelse(cellname_mycluster[["1"]] %in% names(Idents(All.merge)),"cluster1",
ifelse(cellname_mycluster[["3"]] %in% names(Idents(All.merge)),"cluster3","cluster.mixed4_5")))
All.merge@meta.data= All.merge@meta.data%>% mutate(newid2=ifelse(cellname_mycluster[["0"]] %in% colnames(All.merge),"cluster0",
ifelse(cellname_mycluster[["1"]] %in% colnames(All.merge),"cluster1",
ifelse(cellname_mycluster[["3"]] %in% colnames(All.merge),"cluster3","mix4_5"))))
All.merge@meta.data$newid2=ifelse(cellname_mycluster[["0"]] %in% colnames(All.merge),"cluster0",
ifelse(cellname_mycluster[["1"]] %in% colnames(All.merge),"cluster1",
ifelse(cellname_mycluster[["3"]] %in% colnames(All.merge),"cluster3","mix4_5")))
All.merge@meta.data$newid2=ifelse(cellname_mycluster[["0"]] %in% colnames(All.merge),
ifelse(cellname_mycluster[["1"]] %in% colnames(All.merge),
ifelse(cellname_mycluster[["3"]] %in% colnames(All.merge),"cluster3","mix4_5"),"cluster1"),"cluster0")
All.merge@meta.data$newid2=ifelse(cellname_mycluster[["0"]] %in% rownames(All.merge@meta.data),
ifelse(cellname_mycluster[["1"]] %in% rownames(All.merge@meta.data),
ifelse(cellname_mycluster[["3"]] %in% rownames(All.merge@meta.data),"cluster3","mix4_5"),"cluster1"),"cluster0")
All.merge@meta.data$newid2=ifelse(cellname_mycluster[["0"]] %in% rownames(All.merge@meta.data),"cluster0",
ifelse(cellname_mycluster[["1"]] %in% rownames(All.merge@meta.data),"cluster1",
ifelse(cellname_mycluster[["3"]] %in% rownames(All.merge@meta.data),"cluster3","mix4_5")))
All.merge@meta.data$newid2=ifelse( rownames(All.merge@meta.data)==cellname_mycluster[["0"]],"cluster0",
ifelse(rownames(All.merge@meta.data)==cellname_mycluster[["1"]],"cluster1",
ifelse(rownames(All.merge@meta.data)==cellname_mycluster[["3"]],"cluster3","mix4_5")))
All.merge@meta.data$newid2=ifelse( union(rownames(All.merge@meta.data),cellname_mycluster[["0"]]),"cluster0",
ifelse(union(rownames(All.merge@meta.data),cellname_mycluster[["1"]]),"cluster1",
ifelse(union(rownames(All.merge@meta.data),cellname_mycluster[["3"]]),"cluster3","mix4_5")))
All.merge@meta.data$newid2=ifelse( !is.na(intersect(rownames(All.merge@meta.data),cellname_mycluster[["0"]])),"cluster0",
ifelse(!is.na(intersect(rownames(All.merge@meta.data),cellname_mycluster[["1"]])),"cluster1",
ifelse(!is.na(intersect(rownames(All.merge@meta.data),cellname_mycluster[["3"]])),"cluster3","mix4_5")))
All.merge$cellid=rownames(All.merge@meta.data)
All.merge@meta.data$newid2=ifelse( cellname_mycluster[["0"]] %in% rownames(All.merge@meta.data),"cluster0","mix4_5")
All.merge@meta.data$newid2=ifelse( !is.na(intersect(cellname_mycluster[["0"]],rownames(All.merge@meta.data))),"cluster0","mix4_5")
length(row.names(All.merge@meta.data))
length(cellname_mycluster[["0"]])
ncol(All.merge)-length(cellname_mycluster[["0"]])
rep(NA,times=ncol(All.merge)-length(cellname_mycluster[["0"]]))
union(cellname_mycluster[["0"]],
rep(paste("NA-"),times=ncol(All.merge)-length(cellname_mycluster[["0"]])) )
myrep0=c(rep(paste("NA-"),times=ncol(All.merge)-length(cellname_mycluster[["0"]])))
length(myrep0)
union(myrep0,cellname_mycluster[["0"]])
length(union(cellname_mycluster[["0"]],myrep0)) #并集的相同元素会被合并,最终只显示一个相同的元素
union(cellname_mycluster[["0"]],
seq(1,ncol(All.merge)-length(cellname_mycluster[["0"]]),1)
)
union(cellname_mycluster[["0"]],seq(1,ncol(All.merge)-length(cellname_mycluster[["0"]]),1) ) table(All.merge@meta.data$newid2)
seq(1,4,1)
x <- c(1:4)
y <- c(2:7)
union(x,y)
union(c(1,4532,2,4),c("a","h2",4,2))
rep(2,times=4)
length(union(cellname_mycluster[["0"]],rep(NA,times=ncol(All.merge)-length(cellname_mycluster[["0"]]))))
unique(All.merge@meta.data$newid2)
table(All.merge@meta.data$newid2)
grep(rownames(All.merge@meta.data),cellname_mycluster[["0"]])
union(rownames(All.merge@meta.data),cellname_mycluster[["0"]])
head(All.merge@meta.data)
dim(All.merge)
names(Idents(All.merge))
table(All.merge$newid)
table(colnames(All.merge)==cellname_mycluster[["0"]])
match(c(1,3,5),c(2,3,5,78,9,1))
}
#######成功!终于成功了!ifelse判断的是向量是否符合某个条件 此处 左右两边的向量长度必须相等, 于是我用union构造相同长度的向量 但是union里面的元素都是不重合的,不会重复!!!!!!!!!!
All.merge@meta.data$newid2=ifelse(union(cellname_mycluster[["0"]],seq(1,ncol(All.merge)-length(cellname_mycluster[["0"]]),1) ) %in% rownames(All.merge@meta.data),
ifelse(union(cellname_mycluster[["1"]],seq(1,ncol(All.merge)-length(cellname_mycluster[["1"]]),1) ) %in% rownames(All.merge@meta.data),
ifelse(union(cellname_mycluster[["3"]],seq(1,ncol(All.merge)-length(cellname_mycluster[["3"]]),1) ) %in% rownames(All.merge@meta.data),"cluster3","mix4_5"),"cluster1"),"cluster0")
c(1,34,5,4,2,52,1) %in% c(22,124,1,4,2,34,5,2)
seq(1,10-4,1)
All.merge$newid2=ifelse( Idents(All.merge)=="Monocyte","Monocyte",
ifelse( Idents(All.merge)=="T cell","T cell",
ifelse( Idents(All.merge)=="B cell","B cell",
ifelse( Idents(All.merge)=="Ig-producing B cell","Ig-producing B cell",
ifelse( rownames(All.merge@meta.data) %in% union(cellname_mycluster[["0"]],seq(1,ncol(All.merge)-length(cellname_mycluster[["0"]]),1) ),"cluster0",
ifelse(rownames(All.merge@meta.data) %in% union(cellname_mycluster[["1"]],seq(1,ncol(All.merge)-length(cellname_mycluster[["1"]]),1) ),"cluster1",
ifelse(rownames(All.merge@meta.data) %in% union(cellname_mycluster[["3"]],seq(1,ncol(All.merge)-length(cellname_mycluster[["3"]]),1) ),"cluster3","cluster.mixed4_5")
))))))
table(All.merge$newid2)
table(union(cellname_mycluster[["0"]],seq(1,ncol(All.merge)-length(cellname_mycluster[["0"]]),1) ) %in% rownames(All.merge@meta.data))
table(Idents(All.merge)=="Monocyte")
}
library(CellChat)
library(patchwork)
library(ggplot2)
library(ggalluvial)
library(svglite)
library(Seurat)
library(openxlsx)
library(harmony)
library(dplyr)
getwd()
path="G:/silicosis/sicosis/yll/macrophage/no cluster2/0.3/pure_cluster01345_dotplot"
dir.create(path)
setwd(path)
#https://www.jianshu.com/p/cef5663888ff
marker = read.xlsx("G:/silicosis/sicosis/silicosis_ST/overlapped_map/Rigional and cell markers.xlsx",
sheet = "SingleCell_markers")
marker = read.xlsx("G:/silicosis/sicosis/yll/macrophage/no cluster2/0.3/3_Macrophgage_cluster_markers.xlsx")
load("G:/silicosis/sicosis/silicosis-1122-merge/silicosis_cluster_merge.rds")## 改路径
load("G:/silicosis/sicosis/yll/macrophage/no cluster2/macrophage_clean.rds")
getwd()
DimPlot(subset_data)
Idents(subset_data)=subset_data$RNA_snn_res.0.3 #cluster0 1 3 4 5
DimPlot(subset_data,label = TRUE)
table(subset_data$RNA_snn_res.0.3)
cellname_mycluster=list()
for (cluster in c(0,1,3,4,5)) {
#cluster=0
mycluster=cluster
cellname_mycluster[[paste(cluster)]]=colnames(subset_data[,subset_data$RNA_snn_res.0.3==mycluster])
print(paste('cluster' ,"_",cluster,"====="));print(length(cellname_mycluster[[paste(cluster)]]))
}
names(cellname_mycluster)
FindAllMarkers()
DimPlot(All.merge,label = TRUE, cells.highlight=cellname_mycluster[["0"]])+ggtitle(paste("cluster_","0"))
DimPlot(All.merge,label = TRUE, cells.highlight=cellname_mycluster[["1"]])+ggtitle(paste("cluster_","1"))
DimPlot(All.merge,label = TRUE, cells.highlight=cellname_mycluster[["3"]])+ggtitle(paste("cluster_","3"))
DimPlot(All.merge,label = TRUE, cells.highlight=cellname_mycluster[["4"]])+ggtitle(paste("cluster_","4"))
DimPlot(All.merge,label = TRUE, cells.highlight=cellname_mycluster[["5"]])+ggtitle(paste("cluster_","5"))
length(cellname_mycluster[["0"]])
#测试用
if(1==1){#https://www.biostars.org/p/408922/
#AllCells.combined$IGHMPOS <- WhichCells(AllCells.combined, expression = Ighm > 0)
#https://github.com/satijalab/seurat/issues/2081
#https://www.datasciencemadesimple.com/union-in-r/
#https://github.com/satijalab/seurat/issues/2081
#https://github.com/satijalab/seurat/issues/3079
#测试用的
if(1==1){All.merge$newid=ifelse(cellname_mycluster[["0"]] %in% colnames(All.merge),"cluster0",
ifelse(cellname_mycluster[["1"]] %in% colnames(All.merge),"cluster1",
ifelse(cellname_mycluster[["3"]] %in% colnames(All.merge),"cluster3","mix4_5")))
All.merge$newid=ifelse(cellname_mycluster[["0"]] %in% colnames(All.merge),"cluster0",
ifelse(cellname_mycluster[["1"]] %in% colnames(All.merge),"cluster1",
ifelse(cellname_mycluster[["3"]] %in% colnames(All.merge),"cluster3","mix4_5")))
All.merge$newid= ifelse(match(cellname_mycluster[["0"]],colnames(All.merge)),"cluster0",
ifelse(match(cellname_mycluster[["1"]],colnames(All.merge)),"cluster1",
ifelse(match(cellname_mycluster[["3"]],colnames(All.merge)),"cluster3","cluster.mixed4_5")))
All.merge$newid= ifelse(colnames(All.merge)[match(cellname_mycluster[["0"]],colnames(All.merge))],"cluster0",
ifelse(colnames(All.merge)[match(cellname_mycluster[["1"]],colnames(All.merge))],"cluster1",
ifelse(colnames(All.merge)[match(cellname_mycluster[["3"]],colnames(All.merge))],"cluster3","cluster.mixed4_5")))
All.merge$newid= ifelse(cellname_mycluster[["0"]] %in% names(Idents(All.merge)),"cluster0",
ifelse(cellname_mycluster[["1"]] %in% names(Idents(All.merge)),"cluster1",
ifelse(cellname_mycluster[["3"]] %in% names(Idents(All.merge)),"cluster3","cluster.mixed4_5")))
All.merge@meta.data= All.merge@meta.data%>% mutate(newid2=ifelse(cellname_mycluster[["0"]] %in% colnames(All.merge),"cluster0",
ifelse(cellname_mycluster[["1"]] %in% colnames(All.merge),"cluster1",
ifelse(cellname_mycluster[["3"]] %in% colnames(All.merge),"cluster3","mix4_5"))))
All.merge@meta.data$newid2=ifelse(cellname_mycluster[["0"]] %in% colnames(All.merge),"cluster0",
ifelse(cellname_mycluster[["1"]] %in% colnames(All.merge),"cluster1",
ifelse(cellname_mycluster[["3"]] %in% colnames(All.merge),"cluster3","mix4_5")))
All.merge@meta.data$newid2=ifelse(cellname_mycluster[["0"]] %in% colnames(All.merge),
ifelse(cellname_mycluster[["1"]] %in% colnames(All.merge),
ifelse(cellname_mycluster[["3"]] %in% colnames(All.merge),"cluster3","mix4_5"),"cluster1"),"cluster0")
All.merge@meta.data$newid2=ifelse(cellname_mycluster[["0"]] %in% rownames(All.merge@meta.data),
ifelse(cellname_mycluster[["1"]] %in% rownames(All.merge@meta.data),
ifelse(cellname_mycluster[["3"]] %in% rownames(All.merge@meta.data),"cluster3","mix4_5"),"cluster1"),"cluster0")
All.merge@meta.data$newid2=ifelse(cellname_mycluster[["0"]] %in% rownames(All.merge@meta.data),"cluster0",
ifelse(cellname_mycluster[["1"]] %in% rownames(All.merge@meta.data),"cluster1",
ifelse(cellname_mycluster[["3"]] %in% rownames(All.merge@meta.data),"cluster3","mix4_5")))
All.merge@meta.data$newid2=ifelse( rownames(All.merge@meta.data)==cellname_mycluster[["0"]],"cluster0",
ifelse(rownames(All.merge@meta.data)==cellname_mycluster[["1"]],"cluster1",
ifelse(rownames(All.merge@meta.data)==cellname_mycluster[["3"]],"cluster3","mix4_5")))
All.merge@meta.data$newid2=ifelse( union(rownames(All.merge@meta.data),cellname_mycluster[["0"]]),"cluster0",
ifelse(union(rownames(All.merge@meta.data),cellname_mycluster[["1"]]),"cluster1",
ifelse(union(rownames(All.merge@meta.data),cellname_mycluster[["3"]]),"cluster3","mix4_5")))
All.merge@meta.data$newid2=ifelse( !is.na(intersect(rownames(All.merge@meta.data),cellname_mycluster[["0"]])),"cluster0",
ifelse(!is.na(intersect(rownames(All.merge@meta.data),cellname_mycluster[["1"]])),"cluster1",
ifelse(!is.na(intersect(rownames(All.merge@meta.data),cellname_mycluster[["3"]])),"cluster3","mix4_5")))
All.merge$cellid=rownames(All.merge@meta.data)
All.merge@meta.data$newid2=ifelse( cellname_mycluster[["0"]] %in% rownames(All.merge@meta.data),"cluster0","mix4_5")
All.merge@meta.data$newid2=ifelse( !is.na(intersect(cellname_mycluster[["0"]],rownames(All.merge@meta.data))),"cluster0","mix4_5")
length(row.names(All.merge@meta.data))
length(cellname_mycluster[["0"]])
ncol(All.merge)-length(cellname_mycluster[["0"]])
rep(NA,times=ncol(All.merge)-length(cellname_mycluster[["0"]]))
union(cellname_mycluster[["0"]],
rep(paste("NA-"),times=ncol(All.merge)-length(cellname_mycluster[["0"]])) )
myrep0=c(rep(paste("NA-"),times=ncol(All.merge)-length(cellname_mycluster[["0"]])))
length(myrep0)
union(myrep0,cellname_mycluster[["0"]])
length(union(cellname_mycluster[["0"]],myrep0)) #并集的相同元素会被合并,最终只显示一个相同的元素
union(cellname_mycluster[["0"]],
seq(1,ncol(All.merge)-length(cellname_mycluster[["0"]]),1)
)
union(cellname_mycluster[["0"]],seq(1,ncol(All.merge)-length(cellname_mycluster[["0"]]),1) ) table(All.merge@meta.data$newid2)
seq(1,4,1)
x <- c(1:4)
y <- c(2:7)
union(x,y)
union(c(1,4532,2,4),c("a","h2",4,2))
rep(2,times=4)
length(union(cellname_mycluster[["0"]],rep(NA,times=ncol(All.merge)-length(cellname_mycluster[["0"]]))))
unique(All.merge@meta.data$newid2)
table(All.merge@meta.data$newid2)
grep(rownames(All.merge@meta.data),cellname_mycluster[["0"]])
union(rownames(All.merge@meta.data),cellname_mycluster[["0"]])
head(All.merge@meta.data)
dim(All.merge)
names(Idents(All.merge))
table(All.merge$newid)
table(colnames(All.merge)==cellname_mycluster[["0"]])
match(c(1,3,5),c(2,3,5,78,9,1))
}
#######成功!终于成功了!ifelse判断的是向量是否符合某个条件 此处 左右两边的向量长度必须相等, 于是我用union构造相同长度的向量 但是union里面的元素都是不重合的,不会重复!!!!!!!!!!
All.merge@meta.data$newid2=ifelse(union(cellname_mycluster[["0"]],seq(1,ncol(All.merge)-length(cellname_mycluster[["0"]]),1) ) %in% rownames(All.merge@meta.data),
ifelse(union(cellname_mycluster[["1"]],seq(1,ncol(All.merge)-length(cellname_mycluster[["1"]]),1) ) %in% rownames(All.merge@meta.data),
ifelse(union(cellname_mycluster[["3"]],seq(1,ncol(All.merge)-length(cellname_mycluster[["3"]]),1) ) %in% rownames(All.merge@meta.data),"cluster3","mix4_5"),"cluster1"),"cluster0")
c(1,34,5,4,2,52,1) %in% c(22,124,1,4,2,34,5,2)
seq(1,10-4,1)
All.merge$newid2=ifelse( Idents(All.merge)=="Monocyte","Monocyte",
ifelse( Idents(All.merge)=="T cell","T cell",
ifelse( Idents(All.merge)=="B cell","B cell",
ifelse( Idents(All.merge)=="Ig-producing B cell","Ig-producing B cell",
ifelse( rownames(All.merge@meta.data) %in% union(cellname_mycluster[["0"]],seq(1,ncol(All.merge)-length(cellname_mycluster[["0"]]),1) ),"cluster0",
ifelse(rownames(All.merge@meta.data) %in% union(cellname_mycluster[["1"]],seq(1,ncol(All.merge)-length(cellname_mycluster[["1"]]),1) ),"cluster1",
ifelse(rownames(All.merge@meta.data) %in% union(cellname_mycluster[["3"]],seq(1,ncol(All.merge)-length(cellname_mycluster[["3"]]),1) ),"cluster3","cluster.mixed4_5")
))))))
table(All.merge$newid2)
table(union(cellname_mycluster[["0"]],seq(1,ncol(All.merge)-length(cellname_mycluster[["0"]]),1) ) %in% rownames(All.merge@meta.data))
table(Idents(All.merge)=="Monocyte")
}
Idents(All.merge)=All.merge$cell.type
levels(All.merge)
rownames(All.merge@meta.data[Idents(All.merge)=="Monocyte",])
All.merge@meta.data$new.cluster.idents=ifelse( Idents(All.merge)=="Monocyte","Monocyte",
ifelse( Idents(All.merge)=="T cell","T cell",
ifelse( Idents(All.merge)=="B cell","B cell",
ifelse( Idents(All.merge)=="Ig-producing B cell","Ig-producing B cell",
ifelse( Idents(All.merge)=="Dendritic cell","Dendritic cell",
ifelse( Idents(All.merge)=="Neutrophil","Neutrophil",
ifelse( Idents(All.merge)=="NK cell","NK cell",
ifelse(Idents(All.merge)=="Endothelial cell-1","Endothelial cell-1",
ifelse( Idents(All.merge)=="Endothelial cell-2","Endothelial cell-2",
ifelse( Idents(All.merge)=="Fibroblast","Fibroblast",
ifelse(Idents(All.merge)=="Myofibroblast/vascular smooth muscle cell","Myofibroblast/vascular smooth muscle cell",
ifelse(Idents(All.merge)=="Cycling basal cell","Cycling basal cell",
ifelse(Idents(All.merge)=="Ciliated cell","Ciliated cell",
ifelse(Idents(All.merge)=="Clara cell","Clara cell",
ifelse(Idents(All.merge)=="AT1 cell","AT1 cell",
ifelse(Idents(All.merge)=="AT2 cell-1","AT2 cell-1",
ifelse(Idents(All.merge)=="AT2 cell-2","AT2 cell-2",
ifelse(Idents(All.merge)=="Igha+ AT2 cell","Igha+ AT2 cell",
ifelse( rownames(All.merge@meta.data) %in% union(cellname_mycluster[["0"]],seq(1,ncol(All.merge)-length(cellname_mycluster[["0"]]),1) ),"cluster0",
ifelse(rownames(All.merge@meta.data) %in% union(cellname_mycluster[["1"]],seq(1,ncol(All.merge)-length(cellname_mycluster[["1"]]),1) ),"cluster1",
ifelse(rownames(All.merge@meta.data) %in% union(cellname_mycluster[["3"]],seq(1,ncol(All.merge)-length(cellname_mycluster[["3"]]),1) ),"cluster3","cluster.mixed4_5")
))
))))))))))))))))))
table(All.merge$new.cluster.idents)
table(All.merge$cell.type,All.merge$orig.ident)
Idents(All.merge)=All.merge$new.cluster.idents
markers=FindAllMarkers(All.merge,only.pos = TRUE,min.pct = 0.25)
head(markers)
mymarker=markers %>% filter(cluster %in% c("cluster1",
"cluster.mixed4_5", "cluster2","cluster3")) %>%
group_by(cluster) %>%slice_max(avg_log2FC,n=200) %>%slice_min(p_val,n=180)
cluster_gene_stat=mymarker
cluster_gene_stat=markers[markers$cluster %in% c("cluster1",
"cluster.mixed4_5", "cluster0","cluster3"), ]
head(cluster_gene_stat)
unique(cluster_gene_stat$cluster)
table(cluster_gene_stat$cluster)
cluster_marker=list()
for(i in unique(cluster_gene_stat$cluster)){ #按照p0.05值、flodchange 1标准 获得11个cluster的marker基因 列表
cluster_marker[[paste("cluster", i, sep = "_")]] <- cluster_gene_stat[cluster_gene_stat$avg_log2FC>1
&cluster_gene_stat$p_val<0.001
&cluster_gene_stat$cluster==i, "gene"]
}
head(cluster_marker)
unlist(lapply(cluster_marker, length))
getwd()
#批量做气泡图 Dotplot
for (j in names(cluster_marker)) {#一定要记得是向量 而不是一个数 for j in(1:11) 一定记得加括号!!!1:length(cluster_marker!!!!!!!!!
#j=5
times=(length(cluster_marker[[j]])+10)/10
times
if (floor(times)>=2 ){
for (i in seq(1,330,10)) { #使用循环出图 画图
#i=1
if ((i+9)>(10*floor(times))) {next}
print(cluster_marker[[j]][i:(i+9)])
jpeg(paste0("cluster_",j,"_",paste0(unlist(strsplit(cluster_marker[[j]][i:(i+2)], " "))[seq(1,3,1)],seq='_',collapse = "") , ".jpeg"),height = 9, width = 18, units = 'in', res=300)
p=DotPlot(All.merge,
features = cluster_marker[[j]][i:(i+9)])
print(p)
dev.off()
print(paste0("cluster_",j));print(cluster_marker[[j]][i:(i+9)])
}
print(j)
} else{ #条件一定记得加括号! !!!!!!!!!!!!!!!11!!!!!!!!!!!
print(j); print(cluster_marker[[j]][1:(1+9)])
jpeg(paste0("cluster_",j,"_",paste0(unlist(strsplit(cluster_marker[[j]][1:(1+2)], " "))[seq(1,3,1)],seq='_',collapse = "") , ".jpeg"),height = 9, width = 18, units = 'in', res=300)
p=DotPlot(All.merge,features = cluster_marker[[j]][1:(1+9)])
print(p)
dev.off()
}
}
if(1==1){head(marker)
unique(marker$cluster)
marker$`macroph-1`
names(marker)
marker$`macroph-1`
cellname="macroph-1"
library(Hmisc)
mymarker=na.omit(unique(marker$"macroph-1")) %>% capitalize()
mymarker
length(mymarker) #查看一共有几个基因
class(mymarker)
str(mymarker)}
mymarker=marker %>% filter(p_val==0 &cluster==1) %>% select(gene)
mymarker=marker %>% filter(p_val==0 &cluster==0) %>% select(gene)
mymarker=marker %>% filter(p_val<0.0000000000000000001 &cluster==4) %>% select(gene)
mymarker=marker %>% filter(p_val<0.00000000000001 &cluster==5) %>% select(gene)
mymarker=marker %>% filter(p_val==0 &cluster==3) %>% select(gene)
DotPlot(All.merge,features = mymarker)
DimPlot(All.merge)
mymarker=mymarker$gene[1:15]
macrophage1_gene=as.list(mymarker)
macrophage1_gene=list(mymarker)
macrophage1_gene=list(c("C1qa","C1qb","C1qc"))
macrophage1_gene=list(c("Ccl2","Gpnmb"))
load("G:/silicosis/sicosis/silicosis_ST/yll/0214/harmony_cluster/d_all/silicosis_ST_harmony_SCT_r0.6.rds")
table(Idents(d.all))
Idents(d.all)<-d.all$SCT_snn_res.0.6
table(d.all@meta.data$SCT_snn_res.0.6)
table(d.all@meta.data$SCT_snn_res.0.8,d.all$stim)
table(d.all@meta.data$SCT_snn_res.0.6,d.all$stim)
#对给定的基因集合进行打分
d.all=AddModuleScore(d.all,
features = macrophage1_gene,
name = "cluster4")
#结果保存在这里
colnames(d.all@meta.data)
SpatialFeaturePlot(d.all,features = c("cluster41"),
pt.size.factor = 2.2)+
ggtitle(paste(macrophage1_gene[[1]], collapse = "|"))
d.all@meta.data$"macrophage1_score"
VlnPlot(d.all,features = "macrophage1_gene1")
SpatialDimPlot(d.all, images = "image", label = FALSE
) #sio2_7d
SpatialFeaturePlot(d.all,features = c("macrophage1_gene1"))
SpatialFeaturePlot(NS_7_sct, features = markers[1], slot = "scale.data")+
ggtitle(paste(markers, collapse = "|"))
#ggplot画 ump 和tsne 从seurat中使用addmodule得到的umap 使用ggplot画图
#####
########ggplot画 ump 和tsne 从seurat中使用addmodule得到的umap 使用ggplot画图
load("G:/silicosis/sicosis/silicosis-1122-merge/silicosis_cluster_merge.rds")## 改路径
getwd()
path="G:/silicosis/sicosis/silicosis_ST/yll/0214/harmony_cluster/"
dir.create(path)
setwd(path)
library(openxlsx)
inflammatory_gene=read.xlsx("G:/silicosis/sicosis/silicosis_ST/yll/0214/harmony_cluster/HALLMARK_INFLAMMATORY_RESPONSE.xlsx")
head(inflammatory_gene)
library(Hmisc)
inflammatory_gene=capitalize(tolower(inflammatory_gene$gene_symbol))
inflammatory_gene=list(inflammatory_gene)
#对给定的基因集合进行打分
All.merge=AddModuleScore(All.merge,
features = inflammatory_gene,
name = "inflammatory_gene")
#结果保存在这里
colnames(All.merge@meta.data)
SpatialFeaturePlot(d.all,features = c("cluster41"),
pt.size.factor = 2.2)+
ggtitle(paste(macrophage1_gene[[1]], collapse = "|"))
dim(All.merge)
DimPlot(All.merge,group.by = "cell.type")
VlnPlot(All.merge,features = "inflammatory_gene1",split.by = "stim")
VlnPlot(All.merge,features = "inflammatory_gene1",group.by = "stim")
FeaturePlot(All.merge,split.by = "stim",features ="inflammatory_gene1",label = TRUE,
cols = c("lightgrey" ,"#DE1F1F")) #cols = c(rgb(220,212,213,180,maxColorValue = 255),rgb(174,27,52,50, maxColorValue = 255))
FeaturePlot(All.merge,split.by = "stim",features ="inflammatory_gene1",
cols = c(rgb(220,212,213,180,maxColorValue = 255),rgb(174,27,52,50, maxColorValue = 255))
)
FeaturePlot(All.merge,split.by = "stim",features ="inflammatory_gene1",label = TRUE)+
scale_color_gradientn(values = seq(0,1,0.2),colours = c('blue','cyan','green','yellow','orange','red'))
FeaturePlot(All.merge,features ="inflammatory_gene1",label = TRUE)+
scale_color_gradientn(values = seq(0,1,0.2),colours = c('blue','cyan','green','yellow','orange','red'))+
facet_wrap(~stim)
#ggplot画 ump 和tsne 从seurat中使用addmodule得到的umap 使用ggplot画图
if(1==1){
#ggplot画 ump 和tsne 从seurat中使用addmodule得到的umap 使用ggplot画图
library(ggplot2)
mydata<- FetchData(All.merge,vars = c("UMAP_1","UMAP_2","inflammatory_gene1","stim"))
head(mydata)
boxplot(mydata$inflammatory_gene1)
a <- ggplot(mydata,aes(x = UMAP_1,y =UMAP_2,colour = inflammatory_gene1))+
geom_point(size = 1)+
scale_color_gradientn(values = seq(-0.2,1,0.2),colours = c('blue','cyan','green','yellow','orange','red'))
a+ theme_bw() + theme(panel.border = element_blank(), panel.grid.major = element_blank(),
panel.grid.minor = element_blank(), axis.line = element_line(colour = "black"))
#####
data<- FetchData(All.merge,vars = c("cell.type","inflammatory_gene1","stim"))
head(data)
p <- ggplot(data,aes(cell.type,inflammatory_gene1))+geom_boxplot()+theme_bw()+RotatedAxis()
p+geom_boxplot()+theme_bw()+RotatedAxis()
p+geom_boxplot()+facet_wrap(~stim) +theme_bw()+RotatedAxis()
##############
data<- FetchData(All.merge,vars = c("cell.type","inflammatory_gene1","stim"))
head(data)
p <- ggplot(data,aes(cell.type,inflammatory_gene1))+geom_boxplot()+theme_bw()+RotatedAxis()
p+geom_boxplot()+theme_bw()+RotatedAxis()
p+geom_boxplot()+facet_grid(~stim) +theme_bw()+RotatedAxis()
##########333成功!
data<- FetchData(All.merge,vars = c("cell.type","inflammatory_gene1","stim"))
head(data)
p <- ggplot(data,aes(x=cell.type,y=inflammatory_gene1,fill=stim))+
geom_boxplot()+theme_bw()+RotatedAxis()
p+geom_boxplot()+theme_bw()+RotatedAxis()
#
mydata<- FetchData(All.merge,vars = c("UMAP_1","UMAP_2","inflammatory_gene1","stim"))
head(mydata)
ggplot(mydata,aes(x = UMAP_1,y =UMAP_2,
colour = inflammatory_gene1))+
geom_point(size = 1)
ggplot(mydata,aes(x = UMAP_1,y =UMAP_2,
colour = inflammatory_gene1))+
geom_point(aes(colour=stim),size = 1)
ggplot(mydata,aes(x = UMAP_1,y =UMAP_2,
colour = inflammatory_gene1))+
geom_point(size = 1)+
facet_wrap(~stim)
######成功
ggplot(mydata,aes(x = UMAP_1,y =UMAP_2,
colour = inflammatory_gene1))+
scale_color_gradientn(values = seq(0,1,0.2),colours = c('blue','cyan','green','yellow','orange','red'))+
geom_point(size = 1)+
facet_wrap(~stim)
#new color#https://stackoverflow.com/questions/18487369/ggplot-set-scale-color-gradientn-manually
ggplot2::ggplot(mydata,aes(x = UMAP_1,y =UMAP_2,
colour = inflammatory_gene1))+
scale_colour_gradientn(colours = c("red","yellow","green","lightblue","darkblue"),
values = c(1.0,0.8,0.6,0.4,0.2,0))+
geom_point(size = 1)+
facet_wrap(~stim)
#
ggplot2::ggplot(mydata,aes(x = UMAP_1,y =UMAP_2,
colour = inflammatory_gene1))+
scale_colour_gradientn(colours = rainbow(15))+
geom_point(size = 1)+
facet_wrap(~stim)
#
library(scales)
ggplot2::ggplot(mydata,aes(x = UMAP_1,y =UMAP_2,
colour = inflammatory_gene1))+
scale_colour_gradient2(
low = muted("blue"),
mid = "white",
high = "red",
# space = "Lab",
#na.value = "grey50",
guide = "colourbar",
aesthetics = "colour"
)+
geom_point(size = 1)+
facet_wrap(~stim)
#https://ggplot2.tidyverse.org/reference/scale_gradient.html
ggplot2::ggplot(mydata,aes(x = UMAP_1,y =UMAP_2,
colour = inflammatory_gene1))+
scale_colour_gradient2(
low = muted("blue"),
mid = "white",
high = "red",
# space = "Lab",
#na.value = "grey50",
guide = "colourbar",
aesthetics = "colour"
)+
geom_point(size = 1)+
facet_wrap(~stim)
#https://ggplot2.tidyverse.org/reference/scale_gradient.html
ggplot2::ggplot(mydata,aes(x = UMAP_1,y =UMAP_2,
colour = inflammatory_gene1))+
scale_colour_gradientn(colours = terrain.colors(10))+
geom_point(size = 1)+
facet_wrap(~stim)
#https://ggplot2.tidyverse.org/reference/scale_gradient.html
ggplot2::ggplot(mydata,aes(x = UMAP_1,y =UMAP_2,
colour = inflammatory_gene1))+
scale_colour_gradient(low = "yellow", high = "red", na.value = NA)+
geom_point(size = 1)+
facet_wrap(~stim)
#
mydata<- FetchData(All.merge,vars = c("UMAP_1","UMAP_2","inflammatory_gene1","stim"))
head(mydata)
dev.off()
a <- ggplot(mydata,aes(x = UMAP_1,y =UMAP_2,
colour = inflammatory_gene1))+
geom_point(size = 1)+
scale_color_gradientn(values = seq(0,1,0.2),colours = c('blue','cyan','green','yellow','orange','red'))
a+ theme_bw() + theme(panel.border = element_blank(), panel.grid.major = element_blank(),
panel.grid.minor = element_blank(), axis.line = element_line(colour = "black"))
#####富集得分 富集分数
head(mydata)
ggplot(mydata,aes(x = UMAP_1,y =UMAP_2,
colour = inflammatory_gene1))+geom_point(size = 1)+
scale_color_gradientn(values = seq(0,1,0.2),colours = c('blue','cyan','green','yellow','orange','red'))+
facet_wrap(~stim)
###https://www.jianshu.com/p/67d2decf5517
library(Seurat)
library(dplyr)
head(All.merge@meta.data)
mydata<- FetchData(All.merge,vars = c("UMAP_1","UMAP_2","inflammatory_gene1","stim","cell.type"))
head(mydata)
class_avg <- mydata %>%
group_by(cell.type) %>%
summarise(
UMAP_1 = median(UMAP_1),
UMAP_2 = median(UMAP_2),
inflammatory_gene1=mean(inflammatory_gene1)
)
head(class_avg)
#https://ggplot2.tidyverse.org/reference/geom_text.html
#https://ggplot2-book.org/annotations.html?q=text#annotations
pdf("inflammatoryscore.pdf",width = 15,height = 10)
p=ggplot(mydata,aes(x = UMAP_1,y =UMAP_2,
colour = inflammatory_gene1))+geom_point(size = 1)+
scale_color_gradientn(values = seq(0,1,0.2),colours = c('blue','cyan','green','yellow','orange','red'))+ #face = c("plain", "bold", "italic")
geom_text(aes(label = cell.type,fontface="plain",family="sans"), data = class_avg,colour = "red")+ # ggrepel::geom_text_repel(aes(label = cell.type), data = class_avg) +#添加标签
theme(text=element_text(family="sans",size=18)) +
facet_wrap(~stim)+
theme(panel.background = element_rect(fill='white', colour='black'),
panel.grid=element_blank(), axis.title = element_text(color='black',
family="Arial",size=18),axis.ticks.length = unit(0.4,"lines"),
axis.ticks = element_line(color='black'),
axis.ticks.margin = unit(0.6,"lines"),
axis.line = element_line(colour = "black"),
axis.title.x=element_text(colour='black', size=18),
axis.title.y=element_text(colour='black', size=18),
axis.text=element_text(colour='black',size=18),
legend.title=element_blank(),
legend.text=element_text(family="Arial", size=18),
legend.key=element_blank())+
theme(plot.title = element_text(size=22,colour = "black",face = "bold")) +
guides(colour = guide_legend(override.aes = list(size=5)))
print(p)
dev.off()
##
ggplot2::ggplot(data = mydata,mapping = aes(x = UMAP_1,y =UMAP_2,colour = inflammatory_gene1,
fill=stim))+
geom_bar(stat = "identity",position = "stack") #stat = "identity"条形的高度表示数据的值,由aes()函数的y参数决定的
ggplot(mydata, aes(x = UMAP_1,y =UMAP_2))+
geom_boxplot(aes(fill = stim),position=position_dodge(0.5),width=0.6)
#https://www.jianshu.com/p/8d65a13dbc88
# 使用plot函数可视化UMAP的结果
head(mydata)
mystim=factor(mydata$stim)
head(mystim)
plot(mydata[,c(1,2)],col=mystim,pch=16,asp = 1,
xlab = "UMAP_1",ylab = "UMAP_2",
main = "A UMAP visualization of the All.merge")
# 添加分隔线
abline(h=0,v=0,lty=2,col="gray")
# 添加图例
legend("topright",title = "Species",inset = 0.01,
legend = unique(mystim),pch=16,
col = unique(mystim))
#######################################
# 使用ggplot2包可视化UMAP降维的结果
library(ggplot2)
head(mydata)
ggplot(mydata,aes(UMAP_1,UMAP_2,color=stim)) +
geom_point() + theme_bw() +
geom_hline(yintercept = 0,lty=2,col="red") +
geom_vline(xintercept = 0,lty=2,col="blue",lwd=1) +
theme(plot.title = element_text(hjust = 0.5)) +
labs(x="UMAP_1",y="UMAP_2",
title = "A UMAP visualization of the iris dataset")
}
